Phylogenetic Tree Database For Body Measurement
Phylogenetic comparative methods pcms are used to study the evolution of various biological species ranging from microorganisms to animals and plants.
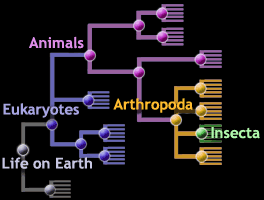
Phylogenetic tree database for body measurement. Thus a phylo4d object connects a phylogenetic tree with a table of trait values and constitutes the basic input for many. The analysis of phylogenetic signal typically involves working with a phylogeny and trait values associated with each tip leaf. Click tree to display phylogentic distribution of a fa. We have demonstrated how to model phylogenetic uncertainty using an empirical prior set of trees derived from the output of bayesian phylogenetic tree estimation programs.
Temporal accuracy of estimation of cultivation antiquity varies among sources but unit of measurements is years ago. Major taxa and level of measurement. Structure images and info for 400 fa. We develop a phylogenetic correlogram for assessing how autocorrelation varies with patristic distance and for judging the appropriateness and effectiveness of an autoregressive model.
The phylobase package hackathon et al. Dataset 1 in txt format containing egg measurements snout vent length and body mass for 259 species of lepidosaurs supplementary table 1 in legendre et al 2020 lepidosaurtreetreesnex. Use of this empirical prior is most attractive because our simulations show that the estimates of regression coefficients are more precise and unbiased for residual variance figure figure8. Phylogenetic tree for dataset 1 in nexus format.
Numerous attributes can be studied across phylogenetic scales tables 1 and 2 which can be defined in terms of clade size clade age tree depth ie time slices through the tree etc. These methods combine trait measurements such as body masses measured in a set of species with the species phylogenetic tree to quantify the traits evolution along the tree. Specifically 866 species of food crops and 901 species of wild progenitors. For a bivariate example we computed estimates of r between body mass and sprint speed using gls ie with no measurement error egls and reml assuming either no phylogenetic relatedness among species a star phylogeny c x c y i or phylogenetic relatedness given by the true phylogeny under brownian motion evolution.
The data points in all three panels represent clades for which the attribute eg diversification rate and the phylogenetic scale eg clade age were calculated. Here we show that current pcms fail to reproduce the. We then revise cheverud et als 1985 evolution 3913351351 autocorrelational model to incorporate greater flexibility in the relation between trait variation and phylogenetic distance. Click name to see species that produce a fa publications and data.